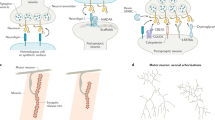
Key Points
-
The organization of neuronal circuits involves a number of processes that require cell–cell recognition and contacts. Cadherins are a family of cell–cell adhesion molecules comprising more than 100 members in vertebrates, which are grouped into subfamilies including classic cadherins, Flamingo/CELSRs and protocadherins, and are thought to have roles in various steps of neuronal cell interactions.
-
N-cadherin and other vertebrate classic cadherins are essential not only for early morphogenesis of neural tissues but also for correct axon migration towards target areas, and for the extension of neuronal dendrites.
-
Drosophila melanogaster N-cadherin (DN-cadherin) has been shown to be crucial for the formation of axonal connections with target neurons in both the visual and olfactory systems, and also for confining dendritic arborizations to specific glomeruli in these systems. The activity of DN-cadherin during the axon targeting seems to be controlled by cytoplasmic proteins including leukocyte antigen-related-receptor protein tyrosine phosphatase (LAR).
-
Flamingo, a seven-pass transmembrane cadherin, is required for the correct targeting of retinal axons in visual circuits in D. melanogaster. A vertebrate homologue of Flamingo, CELSR2, regulates dendritic arbor patterning in the cerebellum, and another homologue, CELSR3, is important for axon tract formation.
-
Some protocadherins, which show a large diversification due to a unique gene organization, seem to be involved in synapse formation and neuronal survival. However, the biological roles of this subfamily remain largely unknown.
-
In conclusion, members of the cadherin superfamily control axon–target recognition and connections, as well as other types of neuronal interactions in a subfamily-specific manner.
Abstract
Neural development and the organization of complex neuronal circuits involve a number of processes that require cell–cell interaction. During these processes, axons choose specific partners for synapse formation and dendrites elaborate arborizations by interacting with other dendrites. The cadherin superfamily is a group of cell surface receptors that is comprised of more than 100 members. The molecular structures and diversity within this family suggest that these molecules regulate the contacts or signalling between neurons in a variety of ways. In this review I discuss the roles of three subfamilies — classic cadherins, Flamingo/CELSRs and protocadherins — in the regulation of neuronal recognition and connectivity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ghysen, A. Dendritic arbors: a tale of living tiles. Curr. Biol. 13, R427–429 (2003).
Chilton, J. K. Molecular mechanisms of axon guidance. Dev. Biol. 292, 13–24 (2006).
Akins, M. R. & Biederer, T. Cell–cell interactions in synaptogenesis. Curr. Opin. Neurobiol. 16, 83–89 (2006).
Sela-Donenfeld, D. & Wilkinson, D. G. Eph receptors: two ways to sharpen boundaries. Curr. Biol. 15, R210–R212 (2005).
Ahimou, F., Mok, L. P., Bardot, B. & Wesley, C. The adhesion force of Notch with Delta and the rate of Notch signaling. J. Cell Biol. 167, 1217–1229 (2004).
Nguyen, T. & Sudhof, T. C. Binding properties of neuroligin 1 and neurexin 1β reveal function as heterophilic cell adhesion molecules. J. Biol. Chem. 272, 26032–26039 (1997).
Tepass, U., Truong, K., Godt, D., Ikura, M. & Peifer, M. Cadherins in embryonic and neural morphogenesis. Nature Rev. Mol. Cell Biol. 1, 91–100 (2000).
Redies, C., Vanhalst, K. & Roy, F. δ-Protocadherins: unique structures and functions. Cell. Mol. Life Sci. 62, 2840–2852 (2005).
Overduin, M. et al. Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science 267, 386–389 (1995).
Shapiro, L. et al. Structural basis of cell–cell adhesion by cadherins. Nature 374, 327–337 (1995).
Takeichi, M. & Abe, K. Synaptic contact dynamics controlled by cadherin and catenins. Trends Cell Biol. 15, 216–221 (2005).
Takeichi, M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science 251, 1451–1455 (1991).
Wheelock, M. J. & Johnson, K. R. Cadherin-mediated cellular signaling. Curr. Opin. Cell Biol. 15, 509–514 (2003).
Drees, F., Pokutta, S., Yamada, S., Nelson, W. J. & Weis, W. I. α-catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell 123, 903–915 (2005).
Rimm, D. L., Koslov, E. R., Kebriaei, P., Cianci, C. D. & Morrow, J. S. α1(E)-catenin is an actin-binding and-bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl Acad. Sci. USA 92, 8813–8817 (1995).
Shimoyama, Y., Tsujimoto, G., Kitajima, M. & Natori, M. Identification of three human type-II classic cadherins and frequent heterophilic interactions between different subclasses of type-II classic cadherins. Biochem. J. 349, 159–167 (2000).
Patel, S. D. et al. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell 124, 1255–1268 (2006).
Inoue, T., Tanaka, T., Suzuki, S. C. & Takeichi, M. Cadherin-6 in the developing mouse brain: expression along restricted connection systems and synaptic localization suggest a potential role in neuronal circuitry. Dev. Dyn. 211, 338–351 (1998).
Suzuki, S. C., Inoue, T., Kimura, Y., Tanaka, T. & Takeichi, M. Neuronal circuits are subdivided by differential expression of type-II classic cadherins in postnatal mouse brains. Mol. Cell. Neurosci. 9, 433–447 (1997).
Cox, E. A., Tuskey, C. & Hardin, J. Cell adhesion receptors in C. elegans. J. Cell Sci. 117, 1867–1870 (2004).
Cox, E. A. & Hardin, J. Sticky worms: adhesion complexes in C. elegans. J. Cell Sci. 117, 1885–1897 (2004).
Tepass, U. Genetic analysis of cadherin function in animal morphogenesis. Curr. Opin. Cell Biol. 11, 540–548 (1999).
Iwai, Y. et al. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron 19, 77–89 (1997). The first paper to have identified D. melanogaster N-cadherin, demonstrating various defects in axon patterning and migration in D N-cadherin-null embryos.
Hatta, K. & Takeichi, M. Expression of N-cadherin adhesion molecules associated with early morphogenetic events in chick development. Nature 320, 447–449 (1986).
Hatta, K., Takagi, S., Fujisawa, H. & Takeichi, M. Spatial and temporal expression pattern of N-cadherin cell adhesion molecules correlated with morphogenetic processes of chicken embryos. Dev. Biol. 120, 215–227 (1987).
Luo, Y. et al. Rescuing the N-cadherin knockout by cardiac-specific expression of N- or E-cadherin. Development 128, 459–469 (2001).
Radice, G. L. et al. Developmental defects in mouse embryos lacking N-cadherin. Dev. Biol. 181, 64–78 (1997).
Erdmann, B., Kirsch, F. P., Rathjen, F. G. & More, M. I. N-cadherin is essential for retinal lamination in the zebrafish. Dev. Dyn. 226, 570–577 (2003).
Malicki, J., Jo, H. & Pujic, Z. Zebrafish N-cadherin, encoded by the glass onion locus, plays an essential role in retinal patterning. Dev. Biol. 259, 95–108 (2003).
Masai, I. et al. N-cadherin mediates retinal lamination, maintenance of forebrain compartments and patterning of retinal neurites. Development 130, 2479–2494 (2003).
Lele, Z. et al. parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development 129, 3281–3294 (2002).
Matsunaga, M., Hatta, K. & Takeichi, M. Role of N-cadherin cell adhesion molecules in the histogenesis of neural retina. Neuron 1, 289–295 (1988).
Inoue, A. & Sanes, J. R. Lamina-specific connectivity in the brain: regulation by N-cadherin, neurotrophins, and glycoconjugates. Science 276, 1428–1431 (1997).
Treubert-Zimmermann, U., Heyers, D. & Redies, C. Targeting axons to specific fiber tracts in vivo by altering cadherin expression. J. Neurosci. 22, 7617–7626 (2002).
Manabe, T. et al. Loss of cadherin-11 adhesion receptor enhances plastic changes in hippocampal synapses and modifies behavioral responses. Mol. Cell. Neurosci. 15, 534–546 (2000).
Kintner, C. Regulation of embryonic cell adhesion by the cadherin cytoplasmic domain. Cell 69, 225–236 (1992).
Fujimori, T. & Takeichi, M. Disruption of epithelial cell–cell adhesion by exogenous expression of a mutated nonfunctional N-cadherin. Mol. Biol. Cell 4, 37–47 (1993).
Hirano, S., Kimoto, N., Shimoyama, Y., Hirohashi, S. & Takeichi, M. Identification of a neural α-catenin as a key regulator of cadherin function and multicellular organization. Cell 70, 293–301 (1992).
Riehl, R. et al. Cadherin function is required for axon outgrowth in retinal ganglion cells in vivo. Neuron 17, 837–848 (1996).
Tanabe, K. et al. Cadherin is required for dendritic morphogenesis and synaptic terminal organization of retinal horizontal cells. Development 133, 4085–4096 (2006). Cadherin activities are shown to be required for the normal extension of horizontal cell dendrites in the retina. Their synaptic formation with photoreceptors is also impaired.
Taniguchi, H., Kawauchi, D., Nishida, K. & Murakami, F. Classic cadherins regulate tangential migration of precerebellar neurons in the caudal hindbrain. Development 133, 1923–1931 (2006).
Uemura, M. & Takeichi, M. αN-catenin deficiency causes defects in axon migration and nuclear organization in restricted regions of the mouse brain. Dev. Dyn. 235, 2559–2566 (2006).
Iwai, Y. et al. DN-cadherin is required for spatial arrangement of nerve terminals and ultrastructural organization of synapses. Mol. Cell. Neurosci. 19, 375–388 (2002).
Clandinin, T. R. & Zipursky, S. L. Making connections in the fly visual system. Neuron 35, 827–841 (2002).
Meinertzhagen, I. A. & O'Neil, S. D. Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J. Comp. Neurol. 305, 232–263 (1991).
Lee, C. H., Herman, T., Clandinin, T. R., Lee, R. & Zipursky, S. L. N-cadherin regulates target specificity in the Drosophila visual system. Neuron 30, 437–450 (2001).
Prakash, S., Caldwell, J. C., Eberl, D. F. & Clandinin, T. R. Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nature Neurosci. 8, 443–450 (2005). Describes precise analyses of the role of D N-cadherin during the targeting processes of R1–R6 axons in the medulla, and demonstrates that both the R axons and their targets require this cadherin to establish synaptic connections.
Ting, C. Y. et al. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development 132, 953–963 (2005).
Nern, A. et al. An isoform-specific allele of Drosophila N-cadherin disrupts a late step of R7 targeting. Proc. Natl Acad. Sci. USA 102, 12944–12949 (2005).
Hummel, T. & Zipursky, S. L. Afferent induction of olfactory glomeruli requires N-cadherin. Neuron 42, 77–88 (2004). D N-cadherin is shown to be essential for the olfactory receptor neuron innervation of the glomeruli in the antennal lobe.
Zhu, H. & Luo, L. Diverse functions of N-cadherin in dendritic and axonal terminal arborization of olfactory projection neurons. Neuron 42, 63–75 (2004). Demonstrates that dendritic branches of the second-order projection neurons forming a glomerulus overspread into neighbouring glomeruli in the absence of DN-cadherin.
Dunah, A. W. et al. LAR receptor protein tyrosine phosphatases in the development and maintenance of excitatory synapses. Nature Neurosci. 8, 458–467 (2005).
Kypta, R. M., Su, H. & Reichardt, L. F. Association between a transmembrane protein tyrosine phosphatase and the cadherin-catenin complex. J. Cell Biol. 134, 1519–1529 (1996).
Clandinin, T. R. et al. Drosophila LAR regulates R1–R6 and R7 target specificity in the visual system. Neuron 32, 237–248 (2001).
Maurel-Zaffran, C., Suzuki, T., Gahmon, G., Treisman, J. E. & Dickson, B. J. Cell-autonomous and-nonautonomous functions of LAR in R7 photoreceptor axon targeting. Neuron 32, 225–235 (2001).
Choe, K. M., Prakash, S., Bright, A. & Clandinin, T. R. Liprin-α is required for photoreceptor target selection in Drosophila. Proc. Natl Acad. Sci. USA 103, 11601–11606 (2006).
Hofmeyer, K., Maurel-Zaffran, C., Sink, H. & Treisman, J. E. Liprin-α has LAR-independent functions in R7 photoreceptor axon targeting. Proc. Natl Acad. Sci. USA 103, 11595–11600 (2006).
Fukata, M. & Kaibuchi, K. Rho-family GTPases in cadherin-mediated cell–cell adhesion. Nature Rev. Mol. Cell Biol. 2, 887–897 (2001).
Tachibana, K. et al. Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J. Cell Biol. 150, 1161–1176 (2000).
Fabre, S. et al. Prominent role of the Ig-like V domain in trans-interactions of nectins. Nectin3 and nectin 4 bind to the predicted C-C′-C′-D β-strands of the nectin1 V domain. J. Biol. Chem. 277, 27006–27013 (2002).
Togashi, H. et al. Interneurite affinity is regulated by heterophilic nectin interactions in concert with the cadherin machinery. J. Cell Biol. 174, 141–151 (2006). Shows that the trans -interactions between nectin 1 and nectin 3 at axon–dendritic contact sites are essential for recruiting cadherins to these sites, which in turn stabilize the synaptic junctions.
Usui, T. et al. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98, 585–595 (1999).
Saburi, S. & McNeill, H. Organising cells into tissues: new roles for cell adhesion molecules in planar cell polarity. Curr. Opin. Cell Biol. 17, 482–488 (2005).
Formstone, C. J. & Mason, I. Expression of the Celsr/flamingo homologue, c-fmi 1, in the early avian embryo indicates a conserved role in neural tube closure and additional roles in asymmetry and somitogenesis. Dev. Dyn. 232, 408–413 (2005).
Formstone, C. J. & Little, P. F. The flamingo-related mouse Celsr family (Celsr1–3) genes exhibit distinct patterns of expression during embryonic development. Mech. Dev. 109, 91–94 (2001).
Hadjantonakis, A. K., Formstone, C. J. & Little, P. F. mCelsr1 is an evolutionarily conserved seven-pass transmembrane receptor and is expressed during mouse embryonic development. Mech. Dev. 78, 91–95 (1998).
Curtin, J. A. et al. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr. Biol. 13, 1129–1133 (2003).
Gao, F. B., Kohwi, M., Brenman, J. E., Jan, L. Y. & Jan, Y. N. Control of dendritic field formation in Drosophila: the roles of flamingo and competition between homologous neurons. Neuron 28, 91–101 (2000).
Sweeney, N. T., Li, W. & Gao, F. B. Genetic manipulation of single neurons in vivo reveals specific roles of flamingo in neuronal morphogenesis. Dev. Biol. 247, 76–88 (2002).
Lee, R. C. et al. The protocadherin Flamingo is required for axon target selection in the Drosophila visual system. Nature Neurosci. 6, 557–563 (2003). Provides evidence that Flamingo is essential for R1–R6 axons to select particular neurons in the lamina during their connection processes.
Senti, K. A. et al. Flamingo regulates R8 axon–axon and axon–target interactions in the Drosophila visual system. Curr. Biol. 13, 828–832 (2003). Shows that, in Flamingo mutants, R8 axons cannot reach the correct positions in the medulla, and also that their axons cannot maintain a correct space between themselves during migration.
Shima, Y. et al. Differential expression of the seven-pass transmembrane cadherin genes Celsr1–3 and distribution of the Celsr2 protein during mouse development. Dev. Dyn. 223, 321–332 (2002).
Tissir, F., De-Backer, O., Goffinet, A. M. & Lambert de Rouvroit, C. Developmental expression profiles of Celsr (Flamingo) genes in the mouse. Mech. Dev. 112, 157–160 (2002).
Shima, Y., Kengaku, M., Hirano, T., Takeichi, M. & Uemura, T. Regulation of dendritic maintenance and growth by a mammalian 7-pass transmembrane cadherin. Dev. Cell 7, 205–216 (2004). The first paper to have examined the role of CELSR2 in neural tissues, showing that dendritic branches of Purkinje cells retract when the expression of this protein is knocked down by RNAi methods.
Tissir, F., Bar, I., Jossin, Y., De Backer, O. & Goffinet, A. M. Protocadherin Celsr3 is crucial in axonal tract development. Nature Neurosci. 8, 451–457 (2005). Demonstrated for the first time that genetic deletion of the Celsr3 gene results in serious defects in various axon tracts in the brain.
Tasic, B. et al. Promoter choice determines splice site selection in protocadherin α and γ pre-mRNA splicing. Mol. Cell 10, 21–33 (2002).
Wang, X., Su, H. & Bradley, A. Molecular mechanisms governing Pcdh-γ gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 16, 1890–1905 (2002).
Esumi, S. et al. Monoallelic yet combinatorial expression of variable exons of the protocadherin-α gene cluster in single neurons. Nature Genet. 37, 171–176 (2005).
Vanhalst, K., Kools, P., Staes, K., van Roy, F. & Redies, C. δ-Protocadherins: a gene family expressed differentially in the mouse brain. Cell. Mol. Life Sci. 62, 1247–1259 (2005).
El-Amraoui, A. & Petit, C. Usher I syndrome: unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J. Cell Sci. 118, 4593–4603 (2005).
Frank, M. et al. Differential expression of individual γ-protocadherins during mouse brain development. Mol. Cell. Neurosci. 29, 603–616 (2005).
Phillips, G. R. et al. γ-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J. Neurosci. 23, 5096–5104 (2003).
Wang, X. et al. γ protocadherins are required for survival of spinal interneurons. Neuron 36, 843–854 (2002). The first paper on Pcdh-γ knockout, which shows that this cadherin is required for neuronal survival in the spinal cord.
Weiner, J. A., Wang, X., Tapia, J. C. & Sanes, J. R. γ protocadherins are required for synaptic development in the spinal cord. Proc. Natl Acad. Sci. USA 102, 8–14 (2005).
Haas, I. G., Frank, M., Veron, N. & Kemler, R. Presenilin-dependent processing and nuclear function of γ-protocadherins. J. Biol. Chem. 280, 9313–9319 (2005).
Hambsch, B., Grinevich, V., Seeburg, P. H. & Schwarz, M. K. γ-Protocadherins, presenilin-mediated release of C-terminal fragment promotes locus expression. J. Biol. Chem. 280, 15888–15897 (2005).
Reiss, K. et al. Regulated ADAM10-dependent ectodomain shedding of γ-protocadherin C3 modulates cell–cell adhesion. J. Biol. Chem. 281, 21735–21744 (2006).
Kohmura, N. et al. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron 20, 1137–1151 (1998).
Blank, M., Triana-Baltzer, G. B., Richards, C. S. & Berg, D. K. α-protocadherins are presynaptic and axonal in nicotinic pathways. Mol. Cell. Neurosci. 26, 530–543 (2004).
Tanoue, T. & Takeichi, M. New insights into Fat cadherins. J. Cell Sci. 118, 2347–2353 (2005).
Down, M. et al. Cloning and expression of the large zebrafish protocadherin gene, Fat. Gene Expr. Patterns 5, 483–490 (2005).
Rock, R., Schrauth, S. & Gessler, M. Expression of mouse dchs1, fjx1, and fat-j suggests conservation of the planar cell polarity pathway identified in Drosophila. Dev. Dyn. 234, 747–755 (2005).
Mitsui, K., Nakajima, D., Ohara, O. & Nakayama, M. Mammalian fat3: a large protein that contains multiple cadherin and EGF-like motifs. Biochem. Biophys. Res. Commun. 290, 1260–1266 (2002).
Nakayama, M., Nakajima, D., Yoshimura, R., Endo, Y. & Ohara, O. MEGF1/fat2 proteins containing extraordinarily large extracellular domains are localized to thin parallel fibers of cerebellar granule cells. Mol. Cell. Neurosci. 20, 563–578 (2002).
Tanoue, T. & Takeichi, M. Mammalian Fat1 cadherin regulates actin dynamics and cell–cell contact. J. Cell Biol. 165, 517–528 (2004).
Ciani, L., Patel, A., Allen, N. D. & ffrench-Constant, C. Mice lacking the giant protocadherin mFAT1 exhibit renal slit junction abnormalities and a partially penetrant cyclopia and anophthalmia phenotype. Mol. Cell Biol. 23, 3575–3582 (2003).
Chu, Y. S. et al. Prototypical type I E-cadherin and type II cadherin-7 mediate very distinct adhesiveness through their extracellular domains. J. Biol. Chem. 281, 2901–2910 (2006).
Morante, J. & Desplan, C. Photoreceptor axons play hide and seek. Nature Neurosci. 8, 401–402 (2005).
Mandai, K. et al. Afadin: a novel actin-filament-binding protein with one PDZ domain localized at cadherin-based cell-to-cell adherens junction. J. Cell Biol. 139, 517–528 (1997).
Acknowledgements
I thank T. R. Clandinin, C. Desplan, H. Togashi and T. Usui for providing the original drawings for schematic illustrations; A. Goffinet, Y. Iwai, I. Masai, K. Tanabe and T. Uemura for providing photographs; S. Hirano for data analysis; and S. Ito for her help in preparing figures. Work in the laboratory was supported by the program Grants-in-Aid for Specially Promoted Research of the Ministry of Education, Science, Sports, and Culture of Japan.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Glossary
- Dendritic field
-
The area covered by dendritic arborizations of a neuron.
- Adherens junction
-
Protein complexes that occur at cell–cell junctions. They are composed of the cadherin–catenin complexes, and characterized by accumulation of actin filaments at their cytoplasmic side.
- Neuroepithelial stage
-
The earliest stage in the developing CNS. The neuroepithelium is a layer of cells with epithelium-like morphologies, which give rise to a diverse array of neural cells during development.
- Ommatidium
-
A unit of the compound eye of insects. Each ommatidium contains a cluster of photoreceptor cells and functionally provides the brain with one picture element.
- Lamina
-
Neuropil structure that makes up part of the optic lobe of insects. Out of eight retinal axons, the R1–R6 axons innervate L1–L5 neurons in the lamina. The lamina L1–L5 neurons relay R1–R6 input to the medulla.
- Medulla
-
Neuropil structure that makes up part of the optic lobe of insects. Out of eight retinal axons the R7 and R8 axons innervate the medulla, which also receives input from the lamina L1–L5 neurons.
- Fascicle
-
A slender bundle of nerve fibres.
- Defasciculation
-
The disentanglement of individual axon fibres from a bundle of fibres, called a fascicle or tract, which allows them to migrate in separate directions.
- Glomerulus
-
In the nervous system, an anatomically discrete module that receives input from other neurons.
- Planar cell polarity
-
(PCP). The property of epithelial cells polarizing along the plane of the epithelium.
- Hemisegment
-
The animal body is segmented along the rostrocaudal axis, as seen in the insects. A hemisegment represents half of a symmetrical segment from either side of the body.
- Stereocilia
-
Mechanosensing organelles of hair cells. As hearing sensors, stereocilia are lined up in the Organ of Corti within the cochlea of the inner ear.
Rights and permissions
About this article
Cite this article
Takeichi, M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci 8, 11–20 (2007). https://doi.org/10.1038/nrn2043
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrn2043
This article is cited by
-
Dysregulation of hypoxia-inducible factor 1α in the sympathetic nervous system accelerates diabetic cardiomyopathy
Cardiovascular Diabetology (2023)
-
Functional convergence of genomic and transcriptomic architecture underlies schooling behaviour in a live-bearing fish
Nature Ecology & Evolution (2023)
-
A delay in vesicle endocytosis by a C-terminal fragment of N-cadherin enhances Aβ synaptotoxicity
Cell Death Discovery (2023)
-
N-Cadherin-Functionalized Nanofiber Hydrogel Facilitates Spinal Cord Injury Repair by Building a Favorable Niche for Neural Stem Cells
Advanced Fiber Materials (2023)
-
Proteomic characterisation of the summer–winter transition in Apis mellifera
Apidologie (2022)