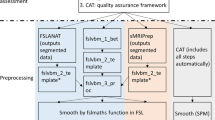
Abstract
Voxel-based morphometry (VBM) has been proven capable of capturing cerebral gray matter asymmetries with a high (voxel-wise) regional specificity. However, a standardized reference on how to conduct voxel-wise asymmetry analyses is missing. This protocol provides the scientific community with a carefully developed guide describing, in 12 distinct steps, how to take structural images from data pre-processing, via statistical analysis, to the final interpretation of the significance maps. Key adaptations compared with the standard VBM workflow involve establishing a voxel-wise hemispheric correspondence, capturing the direction and degree of asymmetry and preventing a blurring of information across hemispheres. The workflow incorporates the most recent methodological developments, including high-dimensional spatial normalization and partial volume estimations. Although the protocol is primarily designed to enable relatively inexperienced users to conduct a voxel-based asymmetry analysis on their own, it may also be useful to experienced users who wish to efficiently adapt their existing scripts or pipelines.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Toga, A.W., Narr, K.L., Thompson, P.M. & Luders, E. Brain Asymmetry: Evolution. in Encyclopedia of Neuroscience Vol. 2 (ed. Squire, L.R.) 303–311 (Academic Press, 2009).
Toga, A.W. & Thompson, P.M. Mapping brain asymmetry. Nat. Rev. Neurosci. 4, 37–48 (2003).
Jancke, L. & Steinmetz, H. Anatomical brain asymmetries and their relevance for functional asymmetries. in The Asymmetrical Brain (eds. Hugdahl, K. & Davidson, R.J.) 187–230 (The MIT Press, 2003).
Luders, E., Gaser, C., Jancke, L. & Schlaug, G. A voxel-based approach to gray matter asymmetries. Neuroimage 22, 656–664 (2004).
Takao, H. et al. Gray and white matter asymmetries in healthy individuals aged 21–29 years: a voxel-based morphometry and diffusion tensor imaging study. Hum. Brain Mapp. 32, 1762–1773 (2011).
Good, C.D. et al. Cerebral asymmetry and the effects of sex and handedness on brain structure: a voxel-based morphometric analysis of 465 normal adult human brains. Neuroimage 14, 685–700 (2001).
Dorsaint-Pierre, R. et al. Asymmetries of the planum temporale and Heschl's gyrus: relationship to language lateralization. Brain 129, 1164–1176 (2006).
Watkins, K.E. et al. Structural asymmetries in the human brain: a voxel-based statistical analysis of 142 MRI scans. Cereb. Cortex 11, 868–877 (2001).
Kurth, F., MacKenzie-Graham, A., Toga, A.W. & Luders, E. Shifting brain asymmetry: the link between meditation and structural lateralization. Soc. Cogn. Affect. Neurosci. doi.org/10.1093/scan/nsu029 (17 March 2014).
Ashburner, J. & Friston, K.J. Voxel-based morphometry: the methods. Neuroimage 11, 805–821 (2000).
Ashburner, J. & Friston, K.J. Why voxel-based morphometry should be used. Neuroimage 14, 1238–1243 (2001).
Ashburner, J. A fast diffeomorphic image registration algorithm. Neuroimage 38, 95–113 (2007).
Ashburner, J. & Friston, K. Voxel-Based Morphometry. in Statistical Parametric Mapping: the Analysis of Functional Brain Images (eds. Friston, K. et al.) 92–100 (Elsevier, 2007).
Tohka, J., Zijdenbos, A. & Evans, A. Fast and robust parameter estimation for statistical partial volume models in brain MRI. Neuroimage 23, 84–97 (2004).
Rajapakse, J.C., Giedd, J.N. & Rapoport, J.L. Statistical approach to segmentation of single-channel cerebral MR images. IEEE Trans. Med. Imaging 16, 176–186 (1997).
Manjon, J.V., Coupe, P., Marti-Bonmati, L., Collins, D.L. & Robles, M. Adaptive non-local means denoising of MR images with spatially varying noise levels. J. Magn. Reson. Imaging 31, 192–203 (2010).
Luders, E., Kurth, F., Toga, A.W., Narr, K.L. & Gaser, C. Meditation effects within the hippocampal complex revealed by voxel-based morphometry and cytoarchitectonic probabilistic mapping. Front. Psychol. 4, 398 (2013).
Wilke, M. & Lidzba, K. LI-tool: a new toolbox to assess lateralization in functional MR-data. J. Neurosci. Methods 163, 128–136 (2007).
Stadler, A., Schima, W., Ba-Ssalamah, A., Kettenbach, J. & Eisenhuber, E. Artifacts in body MR imaging: their appearance and how to eliminate them. Eur. Radiol. 17, 1242–1255 (2007).
Graves, M.J. & Mitchell, D.G. Body MRI artifacts in clinical practice: a physicist's and radiologist's perspective. J. Magn. Reson. Imaging 38, 269–287 (2013).
Rex, D.E. et al. A meta-algorithm for brain extraction in MRI. Neuroimage 23, 625–637 (2004).
Dice, L.R. Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945).
Van Leemput, K., Maes, F., Vandermeulen, D. & Suetens, P. Automated model-based tissue classification of MR images of the brain. IEEE Trans. Med. Imaging 18, 897–908 (1999).
Radua, J., Canales-Rodriguez, E.J., Pomarol-Clotet, E. & Salvador, R. Validity of modulation and optimal settings for advanced voxel-based morphometry. Neuroimage 86, 81–90 (2014).
Ashburner, J. & Friston, K.J. Unified segmentation. Neuroimage 26, 839–851 (2005).
Klein, A. et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 46, 786–802 (2009).
Rohlfing, T. Image similarity and tissue overlaps as surrogates for image registration accuracy: widely used but unreliable. IEEE Trans. Med. Imaging 31, 153–163 (2012).
Friston, K.J., Holmes, A., Poline, J.B., Price, C.J. & Frith, C.D. Detecting activations in PET and fMRI: levels of inference and power. Neuroimage 4, 223–235 (1996).
Hayasaka, S., Phan, K.L., Liberzon, I., Worsley, K.J. & Nichols, T.E. Nonstationary cluster-size inference with random field and permutation methods. Neuroimage 22, 676–687 (2004).
Smith, S.M. & Nichols, T.E. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage 44, 83–98 (2009).
Acknowledgements
This work was supported by the German Ministry of Education and Research (BMBF grant no. 01EV0709 to C.G.).
Author information
Authors and Affiliations
Contributions
F.K. and E.L. developed and designed the protocol and experiments and drafted the manuscript; C.G. developed and wrote the VBM8 tool and provided methodological guidance and feedback; F.K., E.L. and C.G. finalized the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Tissue segmentation.
Shown are examples for successful as well as failed tissue segmentations. The top left displays a successful segmentation of an original T1-weighted image and the two main tissue compartments resulting from the segmentation step: gray matter and white matter. The failed tissue segmentation #1 (top right) revealed a severely distorted gray matter segment and an almost non-existing white matter segment. The problem might be correctable by adjusting the origin of the image (see Troubleshooting Table, Step 1 - Problem 1). The failed tissue segmentation #2 (bottom left) – probably the most likely encountered situation – revealed segments that do not fully comprise the tissue segments (e.g., gray matter is missing in the occipital cortex). The failed tissue segmentation #3 (bottom right) represent a case of severe misclassification of gray and white matter (compare with the segments resulting from the successful tissue segmentation). The cases #2 and #3 might be correctable by adjusting the bias correction (see Troubleshooting Table, Step 1 - Problem 3).
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 and Supplementary Data 1 (PDF 1564 kb)
Supplementary Software 1
MATLAB script called extract.m. (ZIP 2 kb)
Supplementary Software 2
MATLAB script called calculate.m. (ZIP 2 kb)
Rights and permissions
About this article
Cite this article
Kurth, F., Gaser, C. & Luders, E. A 12-step user guide for analyzing voxel-wise gray matter asymmetries in statistical parametric mapping (SPM). Nat Protoc 10, 293–304 (2015). https://doi.org/10.1038/nprot.2015.014
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.014
This article is cited by
-
Visualization of small brain nuclei with a high-spatial resolution, clinically available whole-body PET scanner
Annals of Nuclear Medicine (2024)
-
Altered gray matter volume and functional connectivity in adolescent borderline personality disorder with non-suicidal self-injury behavior
European Child & Adolescent Psychiatry (2024)
-
The effects of environmental factors associated with childhood urbanicity on brain structure and cognition
BMC Psychiatry (2023)
-
Gray matter asymmetry alterations in children and adolescents with comorbid autism spectrum disorder and attention-deficit/hyperactivity disorder
European Child & Adolescent Psychiatry (2023)
-
Hemispheric coupling between structural and functional asymmetries in clinically asymptomatic carotid stenosis with cognitive impairment
Brain Imaging and Behavior (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.