Abstract
Mutations in CHD7, encoding ATP-dependent chromodomain helicase DNA-binding protein 7, in CHARGE syndrome lead to multiple congenital anomalies, including craniofacial malformations, neurological dysfunction and growth delay. Mechanisms underlying the CNS phenotypes remain poorly understood. We found that Chd7 is a direct transcriptional target of oligodendrogenesis-promoting factors Olig2 and Smarca4/Brg1 and is required for proper onset of CNS myelination and remyelination. Genome-occupancy analyses in mice, coupled with transcriptome profiling, revealed that Chd7 interacted with Sox10 and targeted the enhancers of key myelinogenic genes. These analyses identified previously unknown Chd7 targets, including bone formation regulators Osterix (also known as Sp7) and Creb3l2, which are also critical for oligodendrocyte maturation. Thus, Chd7 coordinates with Sox10 to regulate the initiation of myelinogenesis and acts as a molecular nexus of regulatory networks that account for the development of a seemingly diverse array of lineages, including oligodendrocytes and osteoblasts, pointing to previously uncharacterized Chd7 functions in white matter pathogenesis in CHARGE syndrome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Ho, L. & Crabtree, G.R. Chromatin remodelling during development. Nature 463, 474–484 (2010).
Luna-Zurita, L. & Bruneau, B.G. Chromatin modulators as facilitating factors in cellular reprogramming. Curr. Opin. Genet. Dev. 23, 556–561 (2013).
Jakovcevski, M. & Akbarian, S. Epigenetic mechanisms in neurological disease. Nat. Med. 18, 1194–1204 (2012).
Brookes, E. & Shi, Y. Diverse epigenetic mechanisms of human disease. Annu. Rev. Genet. 48, 237–268 (2014).
Martin, D.M. Chromatin remodeling in development and disease: focus on CHD7. PLoS Genet. 6, e1001010 (2010).
Bergman, J.E. et al. CHD7 mutations and CHARGE syndrome: the clinical implications of an expanding phenotype. J. Med. Genet. 48, 334–342 (2011).
Zentner, G.E., Layman, W.S., Martin, D.M. & Scacheri, P.C. Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am. J. Med. Genet. A. 152A, 674–686 (2010).
Engelen, E. et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat. Genet. 43, 607–611 (2011).
Bajpai, R. et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463, 958–962 (2010).
Schnetz, M.P. et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 6, e1001023 (2010).
Bouazoune, K. & Kingston, R.E. Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc. Natl. Acad. Sci. USA 109, 19238–19243 (2012).
Yu, T. et al. Deregulated FGF and homeotic gene expression underlies cerebellar vermis hypoplasia in CHARGE syndrome. eLife 2, e01305 (2013).
Gregory, L.C. et al. Structural pituitary abnormalities associated with CHARGE syndrome. J. Clin. Endocrinol. Metab. 98, E737–E743 (2013).
Liu, L., Yu, T., Wang, L., Mo, X. & Yu, Y. A novel CHD7 mutation in a Chinese patient with CHARGE syndrome. Meta Gene 2, 469–478 (2014).
McKenzie, I.A. et al. Motor skill learning requires active central myelination. Science 346, 318–322 (2014).
Mar, S. & Noetzel, M. Axonal damage in leukodystrophies. Pediatr. Neurol. 42, 239–242 (2010).
Trapp, B.D. et al. Axonal transection in the lesions of multiple sclerosis. N. Engl. J. Med. 338, 278–285 (1998).
Gallo, V. & Deneen, B. Glial development: the crossroads of regeneration and repair in the CNS. Neuron 83, 283–308 (2014).
Zuchero, J.B. & Barres, B.A. Intrinsic and extrinsic control of oligodendrocyte development. Curr. Opin. Neurobiol. 23, 914–920 (2013).
Bercury, K.K. & Macklin, W.B. Dynamics and mechanisms of CNS myelination. Dev. Cell 32, 447–458 (2015).
Nielsen, J.A., Hudson, L.D. & Armstrong, R.C. Nuclear organization in differentiating oligodendrocytes. J. Cell Sci. 115, 4071–4079 (2002).
Yu, Y. et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152, 248–261 (2013).
Kang, S.H., Fukaya, M., Yang, J.K., Rothstein, J.D. & Bergles, D.E. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron 68, 668–681 (2010).
Franklin, R.J. Why does remyelination fail in multiple sclerosis? Nat. Rev. Neurosci. 3, 705–714 (2002).
Stolt, C.C. et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 16, 165–170 (2002).
Emery, B. et al. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell 138, 172–185 (2009).
Swiss, V.A. et al. Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation. PLoS One 6, e18088 (2011).
Bailey, T.L., Williams, N., Misleh, C. & Li, W.W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373 (2006).
Lopez-Anido, C. et al. Differential Sox10 genomic occupancy in myelinating glia. Glia 63, 1897–1914 (2015).
Schuster, N. et al. TGF-beta induces cell death in the oligodendroglial cell line OLI-neu. Glia 40, 95–108 (2002).
Nakashima, K. et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29 (2002).
Saito, A. et al. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat. Cell Biol. 11, 1197–1204 (2009).
Amiel, J. et al. Temporal bone anomaly proposed as a major criteria for diagnosis of CHARGE syndrome. Am. J. Med. Genet. 99, 124–127 (2001).
Friedmann, D.R. et al. Venous malformations of the temporal bone are a common feature in CHARGE syndrome. Laryngoscope 122, 895–900 (2012).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Braitch, M. & Constantinescu, C.S. The role of osteopontin in experimental autoimmune encephalomyelitis (EAE) and multiple sclerosis (MS). Inflamm. Allergy Drug Targets 9, 249–256 (2010).
Comi, C. et al. The impact of osteopontin gene variations on multiple sclerosis development and progression. Clin. Dev. Immunol. 2012, 212893 (2012).
Jongmans, M.C. et al. CHD7 mutations in patients initially diagnosed with Kallmann syndrome--the clinical overlap with CHARGE syndrome. Clin. Genet. 75, 65–71 (2009).
Batsukh, T. et al. CHD8 interacts with CHD7, a protein which is mutated in CHARGE syndrome. Hum. Mol. Genet. 19, 2858–2866 (2010).
Zaret, K.S. & Carroll, J.S. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011).
Voss, T.C. & Hager, G.L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 15, 69–81 (2014).
Zhao, C. et al. Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nat. Commun. (in the press).
Micucci, J.A. et al. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum. Mol. Genet. 23, 434–448 (2014).
Sahlman, J. et al. Premature vertebral endplate ossification and mild disc degeneration in mice after inactivation of one allele belonging to the Col2a1 gene for Type II collagen. Spine 26, 2558–2565 (2001).
Graw, J. Eye development. Curr. Top. Dev. Biol. 90, 343–386 (2010).
Bharti, K. et al. A regulatory loop involving PAX6, MITF, and WNT signaling controls retinal pigment epithelium development. PLoS Genet. 8, e1002757 (2012).
Schimmang, T. Transcription factors that control inner ear development and their potential for transdifferentiation and reprogramming. Hear. Res. 297, 84–90 (2013).
Pauley, S., Lai, E. & Fritzsch, B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev. Dyn. 235, 2470–2482 (2006).
Meyer zum Gottesberge, A.M., Gross, O., Becker-Lendzian, U., Massing, T. & Vogel, W.F. Inner ear defects and hearing loss in mice lacking the collagen receptor DDR1. Lab. Invest. 88, 27–37 (2008).
Huang, J.K. et al. Retinoid X receptor gamma signaling accelerates CNS remyelination. Nat. Neurosci. 14, 45–53 (2011).
Hurd, E.A., Poucher, H.K., Cheng, K., Raphael, Y. & Martin, D.M. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development 137, 3139–3150 (2010).
Xin, M. et al. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J. Neurosci. 25, 1354–1365 (2005).
Sumi-Ichinose, C., Ichinose, H., Metzger, D. & Chambon, P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol. Cell. Biol. 17, 5976–5986 (1997).
Lu, Q.R. et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86 (2002).
Hedtjärn, M., Mallard, C., Arvidsson, P. & Hagberg, H. White matter injury in the immature brain: role of interleukin-18. Neurosci. Lett. 373, 16–20 (2005).
Roughton, K., Boström, M., Kalm, M. & Blomgren, K. Irradiation to the young mouse brain impaired white matter growth more in females than in males. Cell Death Dis. 4, e897 (2013).
Chen, Y. et al. Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat. Protoc. 2, 1044–1051 (2007).
Chan, J.R. et al. NGF controls axonal receptivity to myelination by Schwann cells or oligodendrocytes. Neuron 43, 183–191 (2004).
Trapnell, C., Pachter, L. & Salzberg, S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Acknowledgements
Authors would like to thank Z. Ma, Y. Yang and Z. Tao for technical support. We thank J. Wysocka (Stanford University) for Chd7-expressing vectors and comments, E. Hurlock for suggestions, and P. Wight (University of Arkansas) for the Oli-neu cell line. This study was funded in part by grants from the US National Institutes of Health R01NS072427 and R01NS075243 to Q.R.L., the National Multiple Sclerosis Society (NMSS-4727) to Q.R.L., the Foundation for “l'Aide à la Recherche sur la Sclérose en Plaques” (ARSEP), the National Multiple Sclerosis Society grants RG-4318A1/1 and RG-1501-02851 (to C.P. and Q.R.L.), and the program “Investissements d'avenir” ANR-10- IAIHU-06 to C.P.
Author information
Authors and Affiliations
Contributions
D.H., Q.R.L. and C.P. conceived the studies and designed the experiments. D.H. carried out the studies with Chd7cKO mice and Chd7 target identification and characterization. C.M., M.F. and C.P. performed the studies using Chd7-iKO mice. C.Z. assisted with ChIP-seq experiments. B.K. assisted with histological analysis. J.W. assisted with co-immunoprecipitation experiments. Y.D. and M.F. assisted with LPC-induced demyelination injury. B.V.J. and D.I.C. provided MRI data. D.W. provided human brain samples. X.Z. provided Osterix mutant animals. A.C., H.W., X.H., H.H. and B.Z. provided technical support and intellectual inputs on the manuscript. D.M.M. provided Chd7 floxed mice and advice on the study. D.H., C.P. and Q.R.L. wrote the manuscript with inputs from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
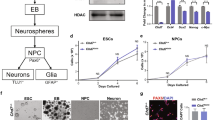
Supplementary Figure 1 The delay of the OL differentiation onset in Chd7 mutant mice
(a) Immunostaining for MBP on the transverse spinal cord sections from control and Chd7cKO mice at e17.5. Nuclei are counterstained with 4,6-diamidino-2-phenylindole (DAPI; blue). Scale bar, 100 μm.
(b) Immunostaining for MBP and Olig2 on the coronal brain sections from control and Chd7cKO mice at P4. Scale bar, 100 μm.
(c) Quantification of number of MBP+ OLs per section from P4 brain and e17.5 spinal cord of control and mutants at the same anatomical levels (mean ± SEM, n = 3; for P4 brain * p = 0.03, t = 3.23; for e17.5 spinal cord ** p = 0.008, t = 4.89; n, number of animals; unpaired two-tailed Student’s t test).
Supplementary Figure 2 Cortical white matter volume and OL numbers are reduced in Chd7 mutant mice
(a) RNA in situ hybridization for Olig1 in P4 and P15 coronal brain (Br) sections from control and Chd7cKO mice. Scale bar, 100 μm.
(b) Quantification of Olig1 ISH density in the corpus callosum of control and Chd7cKO mice at P4 and P15 (mean± SEM; for P4, n = 2, ** p = 0.007, t = 11.359; for P15, n = 3, ** p = 0.0027, t = 6.575; n, number of animals; unpaired two-tailed Student’s t test).
(c) Immunostaining for Sox10 (upper panels) and Olig2 (bottom panels) in the corpus callosum of control and Chd7cKO mice at P9 and P15. Scale bar, 100 μm.
(d) Quantification of Sox10+ and Olig2+ cell density in the control and Chd7cKO corpus callosum at P9 and P15 (mean± SEM; top panels for Sox10: for P9, n = 3, * p = 0.017, t = 3.93; for P15, n = 4, ** p = 0.0027, t = 4.877; bottom panels for Olig2: for P9, n = 3, * p = 0.02, t = 3.74; for P15, n = 4, * p = 0.014, t = 3.38; n, numbers of animals; unpaired two-tailed Student’s t test).
(e) Examples of serial brain sections immunolabeled with MBP from Chd7cKO and control mice at P9. Nuclei are counterstained with DAPI. Scale bar, 1 mm.
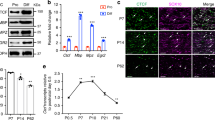
Supplementary Figure 3 OL number and myelination recovery in adult Chd7 mutant mice
(a) Immunolabeling of CC1 on transverse spinal cord sections from Chd7cKO and control mice at postnatal days 4, 7, 15 and 60. Scale bar, 100 μm.
(b) Quantification of CC1+ cells density (top) and CC1+ cell proportion among total Sox10+ cells (bottom) in the white matter region of spinal cords from Chd7cKO and control mice at P1, P4, P7, P15 and P60 (mean ± SEM, n = 3; top panels: for P1, * p = 0.03, t = 3.89; for P4, ** p = 0.0029, t = 6.49; for P7, * p = 0.012, t = 4.36; for P14, * p = 0.045, t = 2.87; for P60, p = 0.71, t = 0.39; n, number of animals; unpaired two-tailed Student’s t test; bottom panels, F(1, 19) = 56.75; Two-way ANOVA test).
(c) Representative electron microscopy images for P60 spinal cord of control and Chd7cKO mice. Scale bar, 2 μm.
(d) Quantification of myelin thickness determined by g-ratio. n = 3 animals of each genotype. No statistical difference of g-ratio between control and mutants.
Supplementary Figure 4 Chd7 deletion in the OL lineage does not affect the development of other neural cell types in the brain
(a) Immunostaining for active cleaved caspase 3 (red) and Olig2 (green) in the corpus callosum from P7 control and Chd7cKO animals. Scale bars, 20 μm.
(b) Immunostaining for GFAP (green) on P7 brain sections from control and Chd7cKO littermates. Nuclei are counterstained with 4,6-diamidino-2-phenylindole (DAPI; blue). Scale bars, 30 μm.
(c) Immunostaining for NeuN (red) on P7 brain sections from control and Chd7cKO littermates. Nuclei are counterstained with DAPI. Scale bars, 100 μm.
Supplementary Figure 5 Impaired OL regeneration in Chd7 mutants during remyelination following LPC-induced demyelination
(a) Representative transverse spinal cord sections from LPC-injected mice stained for Sox10 (upper panels) and Olig2 (bottom panels) at 7 and 14 days post lesion (dpl), as indicated. Scale bar, 100 μm.
(b) Quantification of Sox10+ and Olig2+ in LPC lesion sites at dpl 7 and 14 (mean ± SEM; for Sox10 at dpl 7, n = 4 control and 4 mutant, *p = 0.048, t = 2.47; for Sox10 at dpl 14, n = 4 control and 3 mutant, *p = 0.037, t = 2.82; for Olig2 at dpl 7, n = 4 control and 4 mutant, *p = 0.03, t = 2.81; for Olig2 at dpl 14, n = 4 control and 3 mutant, *p = 0.02, t = 3.33; n, number of animals; unpaired two-tailed Student’s t test).
Supplementary Figure 6 Chd7 is required for brain remyelination
(a,b) Immunofluorescence staining of representative lesions in the corpus callosum from adult (P90) LPC-injected Chd7cKO and control littermates at dpl 7 stained for CC1 (green) & Olig1 (a) or PDGFRα (b) & CC1 (green). Right panels are higher magnification of boxed insets in left panels. Arrows indicate new OLs (CC1high/Olig1high cells) in the remyelinating area. Internal dotted line represents the border of lesion core (almost devoid of OLs) and external dotted line the remyelinating border area. Scale bar, 20 μm.
(c) Quantification of PDGFRα+ cell (OPCs) and CC1 high-expressing cells (differentiating OLs) in LPC lesion sites at Dpl 7 (mean ± SEM, n = 3; for CC1 *** p = 0.0001; t = 15.05; n, number of animals; unpaired two-tailed Student’s t test).
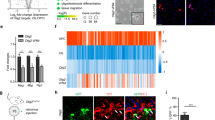
Supplementary Figure 7 Chd7 and Brg1 exhibit distinct binding patterns in differentiating OLs
(a) Venn diagram showing the overlap of Chd7 and Brg1 ChIP-seq peaks in the differentiating OLs.
(b) Heatmap showing the overlapping and unique binding events of Brg1 and Chd7 in differentiating OLs.
(c-e) Representative ChIP-seq genome-browser views of occupancy of Chd7 and Brg1 on Chd7-specific (c), Brg1 (d)-specific and shared (e) binding sites. Genome scale bars: 5 kb.
Supplementary Figure 8 Chd7 activates a cohort of myelination associated genes involved in cytoskeleton organization and lipid metabolism
(a) Venn diagram showing the intersection between Chd7 binding genes and differentially expressed genes in Chd7cKO mutants determined by RNA-Seq. Approximately 144 genes were identified as differentially expressed, Chd7 direct target genes.
(b) Track view of ChIP-Seq density profile for Chd7, H3K27Ac, H3K4me3 and Olig2 centered at selected loci of genes that are functionally important for lipid metabolism (top row) and cytoskeleton organization (bottom row).
(c) Venn diagram showing minimal overlaps of Chd7 genome-occupancy in OLs and ESCs or NSCs, respectively.
(d) Box plot representing Chd7 target gene expression values in log2 FPKM in OLs, astrocytes, neurons, microglial and endothelial cells. Higher expression of Chd7 target genes was observed in OLs. Whiskers show minimum and maximum, box limits are first and third quartile and crosses show average. (n = 704, p < 10–10; n, number of genes with Chd7 occupancy within 5Kb from TSS; Mann–Whitney–Wilcoxon rank test).
Supplementary Figure 9 Chd7 associates with Sox10 at a genome-wide level
(a) MEME motif prediction program indicates that Sox motif rank the top sequence motif enriched at Chd7 ChIP-seq peaks.
(b) Heatmap of Sox10 and Chd7 signal at all Chd7-binding sites. 2 kb upstream and downstream of the anchor sites are plotted.
(c) Sox10 and Chd7 occupancy in Chd7 binding sites are plotted as the average profile of ChIP-seq reads (read density per base pair) around the summit of Chd7 peaks.
Supplementary Figure 10 Chd7 and Sox10 co-occupy on the gene loci of myelination-promoting factors
ChIP-seq genome-browser views of occupancy of Chd7, Sox10, H3K27Ac, H3K4me3 and Olig2 at the same genomic coordinates on indicated myelinogenic genes and Chd7 targeted genes that have not been previously identified. Genome scale bars: 5 kb.
Supplementary Figure 11 Genome occupancy of Chd7 with Sox10 and Olig2
(a) Aggregate plot of the average tag density of Chd7 binding sites at regions occupied by: Chd7 and Sox10 (red), and Chd7 only (green).
(b) Aggregate plot of the average tag density of Chd7 binding sites at regions occupied by: Chd7 and Olig2 (red), and Chd7 only (green).
Supplementary Figure 12 Genome occupancy of Chd7 in the gene loci related to organogenesis
ChIP-seq genome-browser views of occupancy of Chd7, H3K27Ac, H3K4me3 and IgG at the gene loci associated with development and morphogenesis of multiple organs including craniofacial, eye and ear that are affected in CHARGE syndrome patients, such as genes involved in eye development (e.g. Mitf, Rarg, and Pax6), ear development (e.g. Ddr1, Prkra, and Foxg1), and craniofacial and bone development (e.g. Fgfr1/2 and Col2a) in addition to Osterix and Creb3l2. Genome scale bars: 5 kb.
Supplementary Figure 13 Uncropped western blots for Figure 6
Boxed regions were cropped for Figure 6 j and k.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 (PDF 2843 kb)
Supplementary Methods Checklist
(PDF 502 kb)
Rights and permissions
About this article
Cite this article
He, D., Marie, C., Zhao, C. et al. Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nat Neurosci 19, 678–689 (2016). https://doi.org/10.1038/nn.4258
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4258
This article is cited by
-
GBA1 inactivation in oligodendrocytes affects myelination and induces neurodegenerative hallmarks and lipid dyshomeostasis in mice
Molecular Neurodegeneration (2024)
-
ANGPTL2 binds MAG to efficiently enhance oligodendrocyte differentiation
Cell & Bioscience (2023)
-
Systemic and intrinsic functions of ATRX in glial cell fate and CNS myelination in male mice
Nature Communications (2023)
-
Spatiotemporal Dynamics of the Molecular Expression Pattern and Intercellular Interactions in the Glial Scar Response to Spinal Cord Injury
Neuroscience Bulletin (2023)
-
Compound from Magnolia officinalis Ameliorates White Matter Injury by Promoting Oligodendrocyte Maturation in Chronic Cerebral Ischemia Models
Neuroscience Bulletin (2023)