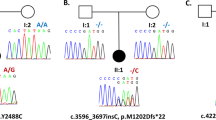
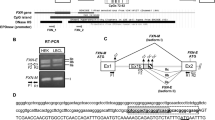
Abstract
DNA methyltransferase 1 (DNMT1) is crucial for maintenance of methylation, gene regulation and chromatin stability1,2,3. DNA mismatch repair, cell cycle regulation in post-mitotic neurons4,5 and neurogenesis6 are influenced by DNA methylation. Here we show that mutations in DNMT1 cause both central and peripheral neurodegeneration in one form of hereditary sensory and autonomic neuropathy with dementia and hearing loss7,8. Exome sequencing led to the identification of DNMT1 mutation c.1484A>G (p.Tyr495Cys) in two American kindreds and one Japanese kindred and a triple nucleotide change, c.1470–1472TCC>ATA (p.Asp490Glu–Pro491Tyr), in one European kindred. All mutations are within the targeting-sequence domain of DNMT1. These mutations cause premature degradation of mutant proteins, reduced methyltransferase activity and impaired heterochromatin binding during the G2 cell cycle phase leading to global hypomethylation and site-specific hypermethylation. Our study shows that DNMT1 mutations cause the aberrant methylation implicated in complex pathogenesis. The discovered DNMT1 mutations provide a new framework for the study of neurodegenerative diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Feng, J. & Fan, G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int. Rev. Neurobiol. 89, 67–84 (2009).
Chen, W.G. et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302, 885–889 (2003).
Tohgi, H. et al. Reduction with age in methylcytosine in the promoter region –224 approximately –101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res. Mol. Brain Res. 70, 288–292 (1999).
Herrup, K. & Yang, Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat. Rev. Neurosci. 8, 368–378 (2007).
Brooks, P.J., Marietta, C. & Goldman, D. DNA mismatch repair and DNA methylation in adult brain neurons. J. Neurosci. 16, 939–945 (1996).
Fan, G. et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J. Neurosci. 21, 788–797 (2001).
Wright, A. & Dyck, P.J. Hereditary sensory neuropathy with sensorineural deafness and early-onset dementia. Neurology 45, 560–562 (1995).
Hojo, K. et al. Hereditary sensory neuropathy with deafness and dementia: a clinical and neuroimaging study. Eur. J. Neurol. 6, 357–361 (1999).
Klein, C.J. & Dyck, P. Hereditary sensory and autonomic neuropathies (HSAN): clinical features pathologic classification, and molecular genetics. in Peripheral Neuropathy, Fourth Edition, Vol. 2 (eds. Peter J. Dyck & P.K. Thomas) 1809–1845 (Philadelphia, USA, 2005).
Dawkins, J.L., Hulme, D.J., Brahmbhatt, S.B., Auer-Grumbach, M. & Nicholson, G.A. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat. Genet. 27, 309–312 (2001).
Rotthier, A. et al. Mutations in the SPTLC2 subunit of serine palmitoyltransferase cause hereditary sensory and autonomic neuropathy type I. Am. J. Hum. Genet. 87, 513–522 (2010).
Guelly, C. et al. Targeted high-throughput sequencing identifies mutations in atlastin-1 as a cause of hereditary sensory neuropathy type I. Am. J. Hum. Genet. 88, 99–105 (2011).
Verhoeven, K. et al. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am. J. Hum. Genet. 72, 722–727 (2003).
Hojo, K., Kawamata, T., Tanaka, C. & Maeda, K. Inflammatory glial activation in the brain of a patient with hereditary sensory neuropathy type 1 with deafness and dementia. Neurosci. Lett. 367, 340–343 (2004).
Fatemi, M., Hermann, A., Pradhan, S. & Jeltsch, A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J. Mol. Biol. 309, 1189–1199 (2001).
Margot, J.B. et al. Structure and function of the mouse DNA methyltransferase gene: Dnmt1 shows a tripartite structure. J. Mol. Biol. 297, 293–300 (2000).
Leonhardt, H., Page, A.W., Weier, H.U. & Bestor, T.H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71, 865–873 (1992).
Fellinger, K., Rothbauer, U., Felle, M., Langst, G. & Leonhardt, H. Dimerization of DNA methyltransferase 1 is mediated by its regulatory domain. J. Cell. Biochem. 106, 521–528 (2009).
Cardoso, M.C. & Leonhardt, H. DNA methyltransferase is actively retained in the cytoplasm during early development. J. Cell Biol. 147, 25–32 (1999).
Easwaran, H.P., Schermelleh, L., Leonhardt, H. & Cardoso, M.C. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 5, 1181–1186 (2004).
Spada, F. et al. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell Biol. 176, 565–571 (2007).
Song, L., James, S.R., Kazim, L. & Karpf, A.R. Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal. Chem. 77, 504–510 (2005).
Chen, T. et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 39, 391–396 (2007).
Brown, K.D. & Robertson, K.D. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat. Genet. 39, 289–290 (2007).
Eden, A., Gaudet, F., Waghmare, A. & Jaenisch, R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300, 455 (2003).
Jones, P.A. & Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3, 415–428 (2002).
Nelson, E.D., Kavalali, E.T. & Monteggia, L.M. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J. Neurosci. 28, 395–406 (2008).
Amir, R.E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 (1999).
Xu, G.L. et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187–191 (1999).
Cao, X. & Sudhof, T.C. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293, 115–120 (2001).
Wijsman, E.M. et al. Evidence for a novel late-onset Alzheimer disease locus on chromosome 19p13.2. Am. J. Hum. Genet. 75, 398–409 (2004).
Tawa, R., Ono, T., Kurishita, A., Okada, S. & Hirose, S. Changes of DNA methylation level during pre- and postnatal periods in mice. Differentiation 45, 44–48 (1990).
Spada, F., Rothbauer, U., Zolghadr, K., Schermelleh, L. & Leonhardt, H. Regulation of DNA methyltransferase 1. Adv. Enzyme Regul. 46, 224–234 (2006).
Golshani, P., Hutnick, L., Schweizer, F. & Fan, G. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Relat. Syst. 3, 227–233 (2005).
Rampon, C. et al. Enrichment induces structural changes and recovery from nonspatial memory deficits in CA1 NMDAR1-knockout mice. Nat. Neurosci. 3, 238–244 (2000).
Acknowledgements
This study was supported by the National Institute of Neurologic Disorders and Strokes (NINDS) K08 (NS065007), NINDS R01 (NS36797) and a previous grant from the Muscular Dystrophy Association. The authors wish to thank the Mayo Center for Translational Science Activities (supported by NIH UL1 RR024150) and the Mayo Clinic Bioinformatics Core and Sequencing Core for their excellent assistance. We would also like to thank the support from the Clinical Core of the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567). D.C.W. and M.S. would like to acknowledge the outstanding clinical contributions of J. Platt. Their portion of this work was supported by US National Institutes of Health grants NS21328 and NS070298.
Author information
Authors and Affiliations
Contributions
C.J.K., D.I.S. and P.J.D. directed the study. C.J.K. wrote the paper. C.J.K., P.J.B.D., G.A.N., S.H., K.H., H.Y., D.C.W., M.S., C.L., L.A.B., G.E.S. and W.J.L. evaluated or collated subject data. E.J.A. did the linkage and haplotype analysis. S.M. and B.B. did the next generation sequencing analysis. C.J.K., C.J.W. and Y.W. did the cell culture and protein expression studies, gene sequencing and protein blot analysis. J.M.C. and A.R.K. did the methylation analysis. M.-V.B. and G.M. did the mutagenesis, bacterial protein expression and structural analysis. J.E.P. provided pathologic analysis of autopsy material.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1–5 and Supplementary Note. (PDF 1346 kb)
Rights and permissions
About this article
Cite this article
Klein, C., Botuyan, MV., Wu, Y. et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet 43, 595–600 (2011). https://doi.org/10.1038/ng.830
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.830
This article is cited by
-
First case of autosomal dominant cerebellar ataxia with deafness and narcolepsy (ADCA-DN) with confirmed DMNT1 gene mutation in Spain. Review of the DMNT1 mutation syndromes
Neurological Sciences (2024)
-
Differential expression of m5C RNA methyltransferase genes NSUN6 and NSUN7 in Alzheimer’s disease and traumatic brain injury
Molecular Neurobiology (2023)
-
Down-regulation of FA-BRCA Pathway in Cervical Carcinoma Gradually Reversed During the Development of Chemo-tolerance: Clinical Implications
Reproductive Sciences (2023)
-
Making sense of the ageing methylome
Nature Reviews Genetics (2022)
-
Germline Abnormalities in DNA Methylation and Histone Modification and Associated Cancer Risk
Current Hematologic Malignancy Reports (2022)